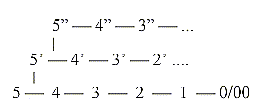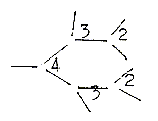|
Some elementary
annotations: 1. 5-6- atoms
in many molecules:
It could
be noted that the number of atoms
(C-N-O-types) on a primary level
in biochemistry is about 5-6, also
the number of dimension degrees
or steps in a dimension chain (transformed
to d-degree 1 giving 5 x 1):
- Carbohydrates as essentially
C5-C6 molecules
- A tetrahedron:
5 atoms.
- A central amino
acid as Glu from α-ketoglutarate
is a C5-molecule.
- Start
of synthesis of fatty acids as C2
+ C3.
- An isoprene molecule:
a C5 molecule.
- Bases: A-base
as 5 x HCN, pyrimidines U-C-T: C4
+ N2, a molecule of 6 ring-forming
atoms. 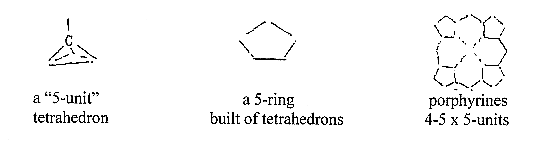
The
PO4 molecule
like the carbon tetrahedron with
one central atom and 4 peripheral
ones are fives, which may be regarded
as still more elementary configurations.
(Why branching
of chains after 3-5 glucose units
in glycogen ?)
2. Forms of these small molecules,
dimensionally interpreted:
-
Tetrahedrons in relation to ring
structures: 3-dimensional.
-
Ring structures in relation to their
own R-groups: 2-dimensional.
- R-groups of the rings in relation
to different kind of bonds: 1-dimensional,
as for instance the H-bonds between
R-groups of the bases in DNA.
It
has been said that a condition for
life is the development from sp3-hybridizations
in tetrahedrons to sp2-hybridization
in planes, dimensionally interpreted
as a step 3 →
2. Such a step towards lower d-degrees
implies increasing number of movements
or kinetic energy according to the
main hypotheses in the model. 3.
Chair- and boat-forms of d-degree
2:
The ring forms as 2-dimensional
illustrate both poles 2a/2b of d-degree
2 in the dimension model, in one
expression described as convex/concave,
this in the "chair" form.
4. Valence numbers and
division in orientation of bonds:
The divisions in directions
of bonds around atoms of different
valences look like illustrations
of d-degree steps: - P-atom
with valence 5 get 4 bonds "outwards",
1 inwards giving one double-bound
oxygen atom. A division 4 + 1.
- C-atom, with valence 4, has its bond directions divided 4 →3
+ 1 in amino acid tetrahedrons as examples. Number 1 represents here
the most differentiated radical line.
(As a guess it's the empty place for an electron
with outward directed spin in the p-orbital of the C-atom that corresponds
to this direction for R-chains of amino acids.)
It's 3 directions is further divided 3 →
2 + 1, where 2 come to represent the division of directions in charge
plus and minus (charge assumed as property in d-degree 2): the polarity
NH3+ <— —> COO- leading to peptide
bonds.
P-group
and amino acid tetrahedrons:
P: 5→
4 + 1 (or 3 + 2, the double-bounded
Oxygen)
C: 4
→
3 + 1
3
→
2 + 1
N: (4)3
→ 2 + 1, (NH3+
2)
O: 2
→ 2 or 1 + 1, (O- 1 +
0 (2 in keto-oxygen, or 1 + 1, e.g. in carbohydrates)
In
ring structures in the codon bases
or carbohydrates and in fatty acids
there is also the division of 4
→2
+ 2 or 2 →
1 +1 for R-groups: 2 forming parts
of the ring or in fatty acids wavy
chain structures giving 2 or 2→1-dimensional
forms. Compare generally waves as
expressions for more motions, lower
d-degrees. - N-atom,
when with valence 3, is in amino
acids for instance or base rings
divided 2 + 1 in directions and
the O-atom with valence 2 has a
still narrower angle between it's
bond directions, 3 in double-bonds,
or 1-1 in water.
5.
Triglycerides divided 2 -1:
On a macromolecule level the
triglycerides undergo a similar
transformation step or division
3 →
2 +1, two hydrophobic fatty acids
in one direction, the 3rd replaced
by a hydrophilic P-group or (later)
other more complex hydrophilic groups.
(The fatty acids already in themselves,
before binding to glycerine, polarized
in hydrophobic and hydrophilic ends.)
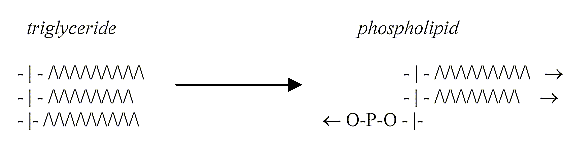
Reevaluating the lipids:
Regarding the structure of phospholipids,
they illustrate in fact most obviously
the 4th d-degree of directions outwards/inwards
in our model - and at the same time
how the d-degree 3 appears through
the polarization in poles 4a --
(3) --- 4b according to the model,
creating the "circular"
cell membrane (with "radial"
canals). Enclosing / excluding,
individualizing units as cells.
6.
Chains and rings as 1-2-dimensional
elements building forms of higher
d-degrees:
Life chemistry
is processes, characterized by increasing
motions, while structures of second
order are of low d-degrees, chains
and rings, principally d-degrees
2 and 1.
In
the dimension model the dimension
chain of structures becomes opposite
in direction to the dimension chain
of motions, with increasing d-degree
in each step outwards in structure:
0/0 ←
1 ←
2 ←
3 ←
4 ← 5
D-degrees
of motions
D-degree
of structure: 5 →
4 →
3 →
2 →
1 →
0/00
| ↓chains (proteins, fatty acids...)
↓
rings (carbohydrates,
nucleic acids...) We
could test to look at the relation
the other way: motional patterns
taking the form of structure d-degree
in the opposite directed dimension
chain: -
protein chains (1) moving "linearly"
(1) in angular steps through 3 dimensions,
when folding to "globular"
proteins, partly also spiralling
(α-helixes),
a 2-3-dimensional motion;
-
carbohydrate chains of rings
spiralling, ~ rotation (2) + pathway
motions (1), also crossed
chains forming surface layers (2-3)
as in cellulose coats of plant cells.
Higher d-degrees
of structure are replaced by motional
patterns as a substantiation of
kinetic energy. 3-dimensional forms
on this level are only created through
motional patterns: It
could perhaps be one way to interpret
the folding of proteins?
(Following
internal bonds between different
parts and the aggregation to dimers
and tetramers possible to regard
as expressions for the 4th d-degree,
e. g. in histones and enzymes?)
7. Displacements in carbon chains
at additions of other ones:
Displacement steps in the connection
between "C-atom-lines"
of the included units seem in many
cases to be a factor in the formation
towards units of higher d-degrees:
In such cases
the added unit don't bind to a C-atom
at the ends of the other but to
the second one or the like:
- One obvious example is when two
δ-aminolevulinic
acids (C5 molecules) combine
to porphobilinogen, where
the 3rd C-atom in one of the molecules
binds to the 4th in the other, leading
to the creation of a 5-ring.
- Another example is the connection
between acetyl~ and dimethylallyl(-P-P)
molecules on the way to
mevalonate and isoprenes.
-
Also the coupling of acetyl~ to
middle C in malonyl~ at the synthesis
of fatty acids.
- The fact that
it is the oxygen group of the second
(an inner) C-atom that is replaced
by an N-group at amination of α-ketoglutarate
to amino acid Glu could illustrate
the same principle?
Such
features could eventually be interpreted
as expression for primary versus
secondary development within a dimension
chain:
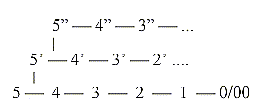
8. Formation
of rings in 5-6 types:
Forms of molecules "of second
order", combined smallest molecules,
are chains and rings, i.g. d-degree
1-2-(3). (Eventually connected with
step 1-2 on the atomic level, the
p-orbital, to which the C-, N-,
O-atoms belong?)
There
seems to be at least 5 quite different
types of ring formations, as if
"every tool" for the purpose
of ring-closing were used. They
are however more or less connected
with the different main classes
of substances and perhaps but not
easily possible to interpret as
formations in different d-degrees
in the model, in a secondary dimension
chain developed within step 2 →←1. ?
The step from
tetrahedrons with central atoms
to ring-formed molecules (cf. sp3
→
sp2) may be regarded
as a projection of centre to anticentre,
to a circumference, as of the 6
edges in the tetrahedron; a displacement
to a new level, a level of lower
d-degree.
According
to general hypotheses here a whole
dimension chain may also close stepwise
towards a ring form through angle
steps if in the same direction.
Figures
below are only vaguely suggested
associations with geometries of
the different d-degrees, surely
debatable. 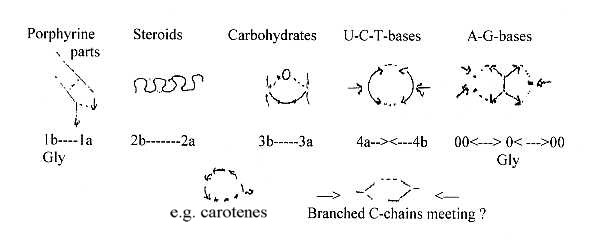
a.
Porphyrine synthesis
with two nearly parallel or anti-parallel
C-chains who through side bonds
form 5-atom rings. b. Synthesis
of steroids where
the wavy chains (of squalene)
give closed rings in both directions,
convex/concave as poles 2a-2b of
d-degree 2. c. The simple
bending of the C-chain (3a) in carbohydrates
bound as through radii (3b) from
an O-atom as centre, a loop formation. d.
The meeting (opposite directions
as 4a--4b) of two bent C- and or
C-N-chains forming "half circles"
of a ring as in the creation of
the pyrimidine rings,U-C-T-bases.
e. Creation of double
rings as in purines
(A-G-bases) from a "radial"
centre (0) (there the amino acid
Gly, a tetrahedron) and a multitude
of small individual molecules meeting
from outside (00) supplementing
and completing the ring forms.
Theoretically 3 steps of bifurcations
can give a 6-atom-ring.
To compare
with 3 polarizations from d-degree
5 to 2.
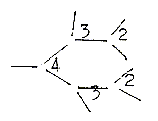
(The interpretation
of the ring forms belong to the
many ambiguities in the dimension
model. A molecular ring may be interpreted
as a surface, d-degree 2, further
as a circumference (pole 3a of d-degree
2) or as a secondary, complex centre
on a superposed level as in the
many ring-formed coenzymes in relation
to the surrounding globular protein
enzymes, Ep.)
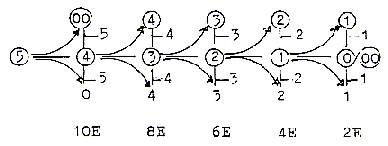
9. Z-numbers of most
elementary molecules in a "10-chain":
Number 10 stepwise polarized,
giving OH-H, O-H2, N-H3, C-H4:
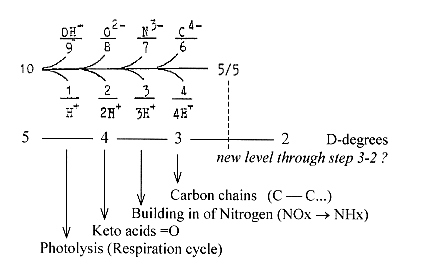
(Abiut
the Citrate cycle: In reality there
seems to be - 5 H from substances/molecules
in the citrate cycle. If 8 H are
gained to the respiration cycle,
3 of them ought to come from outside
during the circle…?) A
level development within step 3-2
. The "10-chain"
may be regarded as a doubled elementary
dimension chain with its two polarizations
5 →
4 + 1, 5 →3
+ 2.
9-1,
7-3 in that case as partitions in
the half steps 5 →
4, 4 →3. H-atoms
= the d-degrees which are lost during
the stepwise process towards lower
d-degrees, translated into movements.
Compare H+ as an essential "force"
or "carrier of force"
in biochemistry.
2
H = "2 E", E for the sum
of poles in respective d-degree,
a kind of energy value in this model.
Total sum of a dimension chain give
sum of poles = 30 "E".
*
|