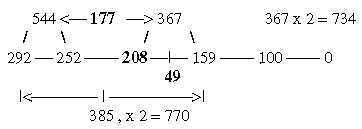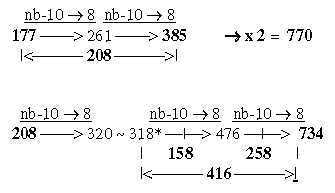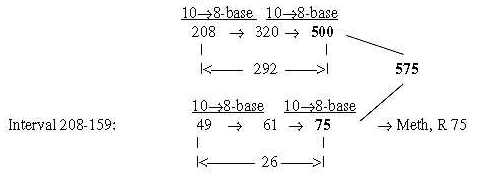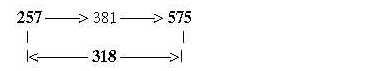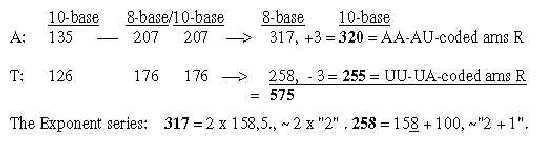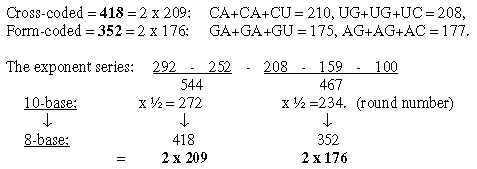|
|
|---|
|
Astronomy
- links:
|
Elements - links:
|
The Genetic Code -
links:
|
Language - links
|
|
17. Transformations between number-base systems (nb-x) Bases ←→
totals of ams − 5 x ES- numbers − Generative production of the 12-groups |
Hence, 4 sets of the 4 bases give the total sum of 24 unbound ams. We find also that 2 x G+C-bases in nb-8 as 768 gives total sum 3276 in nb-6:
The sum of the 4 bases in nb-8 = 752 -/+1:
Fig 17-2. From 752 as sum of ES-numbers 5', 4' and 3' to 2848 in nb-6:
It follows that all operations as multiplications are performed
in nb-10. Indexes for x in nb-x are often used below to shorten
the text. As mentioned above numbers in nb-8 and nb-6 are often
rewritten with figures from nb-10. Another question is how to interpret nb-16 in many examples below If keeping to the thought of x in nb-x as first three numbers in the elementary chain 5' →> 4' →>3' doubled, should nb-16 be regarded as 2 x 4 doubled or 2(5 + 3) doubled?
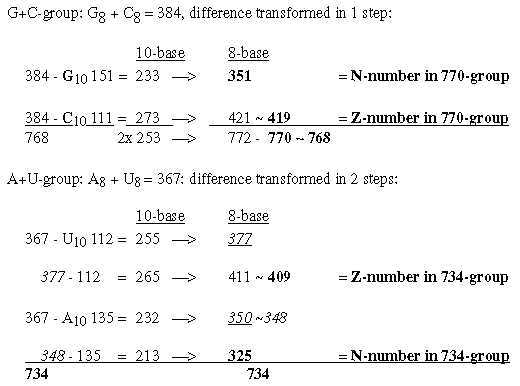
A general feature may be noted: transformation of sums or whole units give larger numbers in lower nb-systems than their parts transformed and summed afterwards. 1.3 Halves of the 12-groups 770 and 734, -/+1 = 384 and 368: Fatty acids, a first annotation here:
Cf. the hexagonal pattern in Table 0 here: fatty acids as a third way to read such a pattern. From the numbers 384 and 368 in nb-10 transformed in two steps to nb-8 we get 2 sets of bases G and A in nb-8, as in opposite direction to the figure above and without C and U:
1.4.1 Four times G+U and A+C to ~ B- and R-chains of total 3276: Sums of R+B-chains together in nb-10: Fig. 17-3 In nb-10 we have groups of ams paired in keto-/amino types: Fig. 17-4
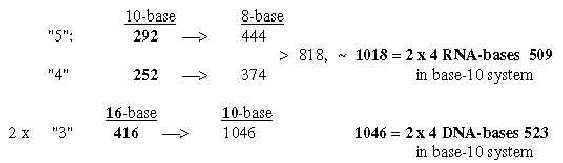
1.4.2 Two sets of bases from ES-numbers 5', 4' and 3':
Number 416 (2 x 3', 208) is the one which added to 544 gives the A-U-group of ams. Cf. that U-base gets replaced by T-base in DNA, a CH2-group added for inward direction to DNA. (It could perhaps be compared with the interpretation of nb-16 as 2 x (3' + 5'), a step backwards from 3' to 5', equivalent with inwards?
2. The bases in the ES-chain: Fig. 17-6 U 160 + C 157 in nb-8 approximate number 2' = 159 in the ES-series,
together 317. These relations could be a reason why G+C-bases get connected with the 12-group 770 of ams in spite of all bases equally represented in this group.
3. 5 times ES-numbers: Fig 17-8: Main
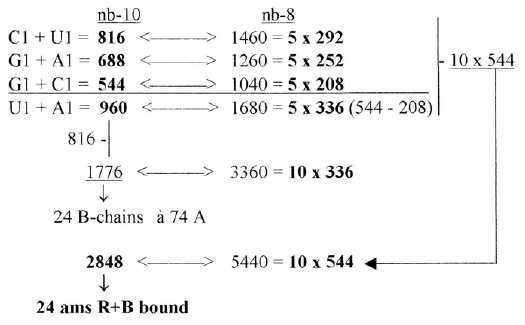
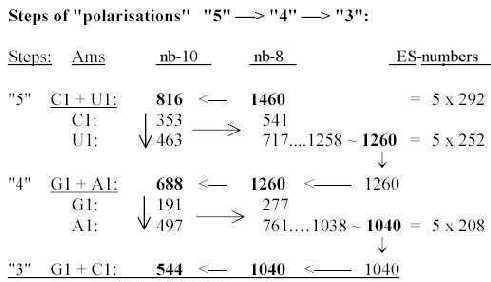
codon groups of ams from 5 times ES-numbers: 816 and 688 is the division of R-chains of total sum 1504 of 24 ams, a division between purine and pyrimidine codon groups, As a division in step 5 - 4 here it precedes the one between complementary pairs G-C and U-A, which are attained from the secondary division of 544 in 336 and 208, a division in step 4 - 3. Note also about 1344, the 24 B-chains bound, included in sum 2848:
These relations seem to support the relevance of both the ES-chain and the thought that nb-transformations could be part of the reference system. Fig 17-9 5 x half of 752, number 688 as an interval: There is also the feature that divisions stepwise as polarizations of numbers 816 in U1 + C1, separately transformed to nb-8 give 1260, next lower level, and this back to nb-10 and divided G1 and A1 gives 1040 in nb-8: Fig 17-10. Stepwise polarization giving next number x5 in Es-chain:
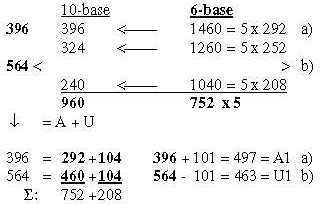
3.3 5 times intervals in the exponent series in nb-8 give ams-groups -/+1: Fig. 17-12 5 x interval in the ES-chain: 3.4 Nb-6: 5 times the ES-numbers 5', 4' 3' in nb-6: Fig..17-13 5 times ES-numbers in nb-6 to 396-564.
3.5 The parts above of 960 in nb-16 gives the total mass of bound ams in nb-8: Fig 17-14 3.6 The numbers U1 + C1 = 816 and G1+ A1 = 688 read in nb-8 rewritten, give in two steps total 24 ams, R + B inbound in nb-8:
4. Generation of the two 12-groups of ams with mixed and non-mixed codons: 4.1 Generative production of sums within 12-groups of ams: 17-15a. The ES-chain, numbers 177 and 208: Cf Table 2,3 in file 02. Numbers 770 and 734 generated from 177 and 208: Fig. 17-15b
Fig 17-15c:
Fig 17-15-d:
Note too that Meth leaves its outer CH3-goup at start of synthesis, (= -15 +1), which gives R-chain = 61, the intermediate number in the figure above. 575 directly from 208 + 49 = 257 in only two steps: Number 75, R-chain of Meth:
4.2 A- and T-bases give the sum 575 of ams with non-mixed codons: Fig. 17-16: A+T How explain the T-base here, a DNA-base giving A in RNA?
4.3 770-group from 4':
Fig 17-17: From halved ES-parts to mixed codon groups
Fig. 17-18:
To Totals and other notable things.
|
© Åsa Wohlin
Individual research
E-mail: a.wohlin@u5d.net
Table
24 amino acids (ams)
R-chains, A, Z, N
Abbreviations
- ways of writing -
Background
model
Files here:
All
these files gathered
in one document, word,
124 p.
All these files in 3 documents, pdf:
Section
I, files 0-11
Section
II, files 12-16
Section
III, files 17-22
Discusssion, References in section III
To
17 short files.
- partly other material
-
The
17 files as one document,
pdf
An
earlier version (2007)
with more material
on the same subject, 73 pages:
Contact:
u5d
Latest updated:
2022-11-12
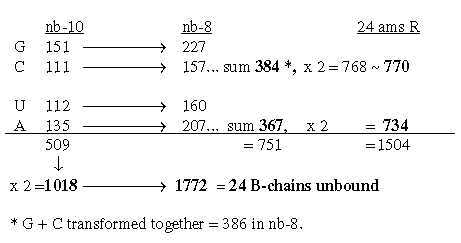
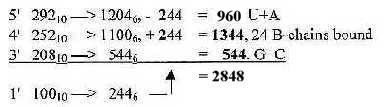 2848 = 24 ams R + B bound
2848 = 24 ams R + B bound Fig Ti-1
Fig Ti-1
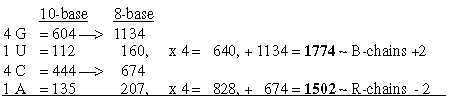
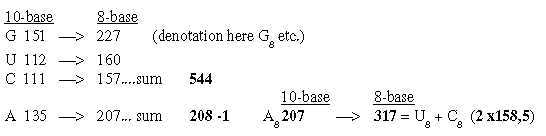
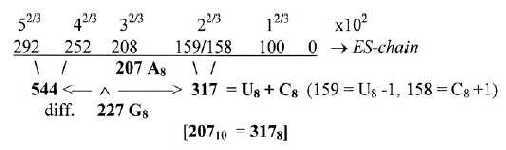
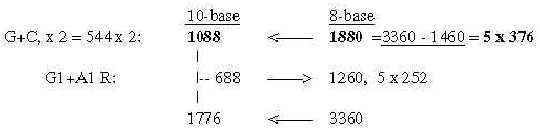
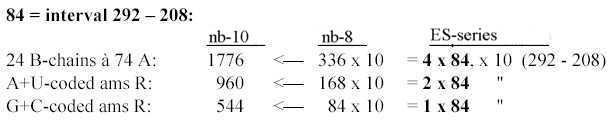
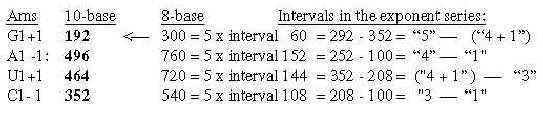
 :
: