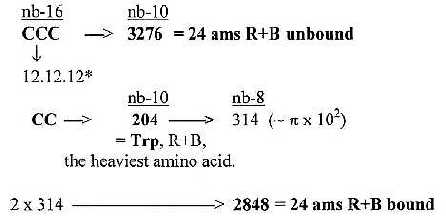
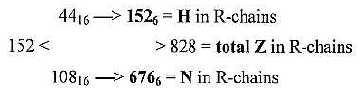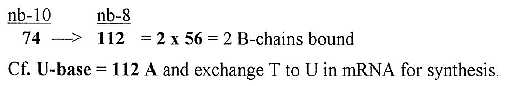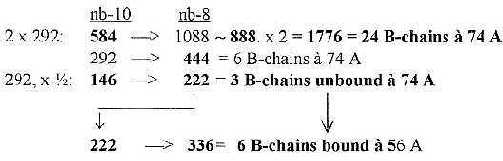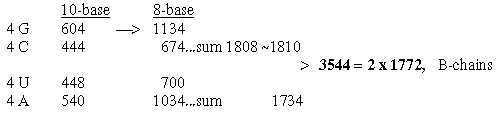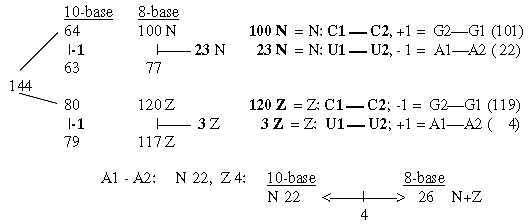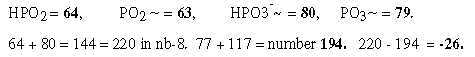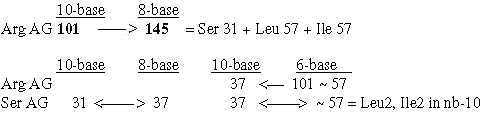1. Total sum R+B-chains of 24 ams unbound = 3276:
3276 is about 1/10 of 215. In nb-16 it's CCC, which
may be transcribed as
12.12.12 = 3072 (3 x 322 = 4 x 768) + 192 + 12:
Fig 18-1: Total
sum of 24 ams R+B:
(2 π x 100:
the bound 24 ams as a closed circle!)
12.12.12: An association goes to carbon 12C and the 3C-molecules
from halved fructose in glycolysis from which first group of ams
derives. Could we eventually read positions of the carbon atoms
as decided and guided by oxygen 16O in some way?! Much
of the process in glycolysis seems to be about a stepwise displacement
of oxygen along the C-C-C-chain.
2. Why 24 ams?
One reason to suspect nb-transformations could be the 4 double-coded
ams, If 20 ams have to be 24, then 4 ams must be repeated (!).
20-10 → 24-8
3. H-atoms, 152 in R-chains: and the total of R 1504:
Number of hydrogen atoms in R-chains was 152 = interval 4'- 1' in
the ES-series.
292 — 252 —208 —— 159 —
100 — 0
———-|<-44-><|——108——>|
This interval is divided 4'-3' = 44 and 3'-1' = 108: Transformed
from nb-16 to nb-6 they give total Z-numbers of R-chains and N-numbers
separately:
Fig 18-2: H-atoms:
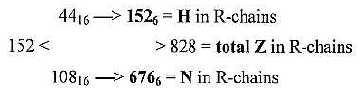
Steps 44 → 152 = + 108
Step 108 → 676 = + 568...This
sum is also = 676 = Z (or N) of atoms C, N. O, S.
Cf. 676 = 262 and the 2x2-chain,file 13.
4. N-numbers in codon-groups of ams may lead to totals of ams:
Fig. 18-3: Neutron numbers to totals

5. Number of C-atoms in R-chains as basis for divisions:
In file 04, para. 3, the ams were ordered after number of C-atoms in their R-chains and their mass summed. This division did not concern codon distribution but seemed related
to the ES-series with certain assumptions.
Here C for carbon. (8 ams with 4 C in R-chains got the sum 584 2 x 292.)
Phe and Tyr are synthesized as 3C- plus 4C-molecules,
hence positioned between 4C- and 3C-.groups. Trp as 3C + 4C + 5C
- 1C. Trp gets its B-chain from Ser, shares codon with Cys and can
brake down to Ala, hence here regarded as "meeting the other
way around", added to the l C group.
Fig. 18-4a: Transformations along the ES-chain as a nxC-chain:
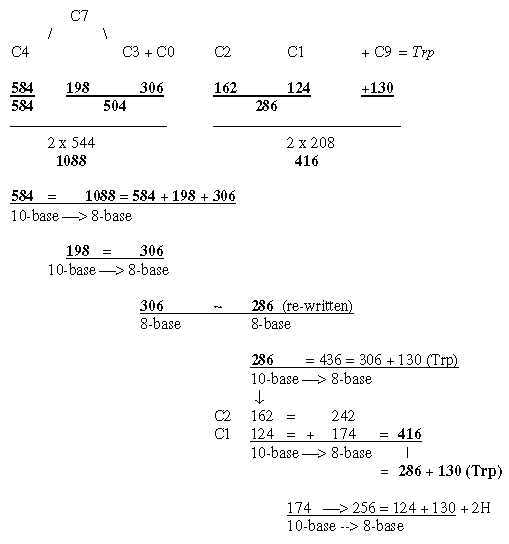
Fig. 18-4b: Cf. triplet sums, file 15, numbers 714 and 792:

Fig. 18-4c: nxC-atoms - three more details:
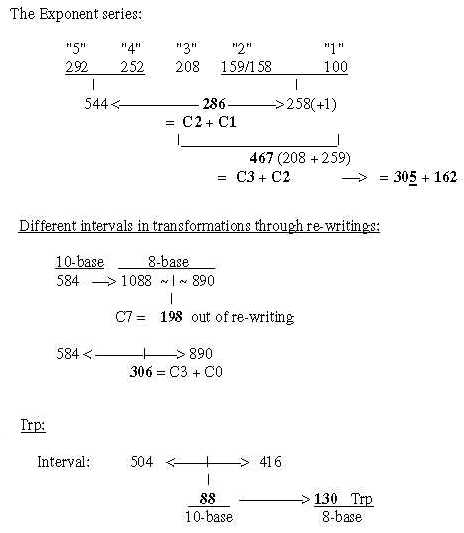
6. B-chains:
6.1 Number 752, sum of first 3 numbers in the ES-chain:
a) 752 from nb-16 to nb-10 gives the total 1772 of
24 B-chains unbound:
292-16 →
658-10
252-16 →
594-10
208-16 → 520-10...
sum 1772, 24 B-chains unbound
Cf. that 752: nb-10 gave 2848 in nb-6, i.e., R+B-chains
of 24 ams bound:
Fig. 18-5:
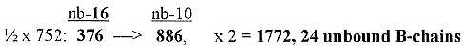
6.2 A single, unbound B-chain = 74:
Two sets of the 4 RNA-bases, sum 1018, gave in nb-8 the
sum of 24 B-chains unbound = 1772. A single unbound B-chain à 74 gives the
sum of 2 bound B-chains.
Fig 18-6: From one unbound
B-chain to two bound ones:
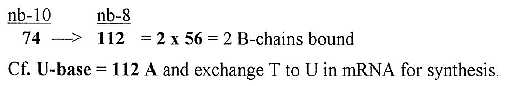
6.3 Halvings of 2 x 5' 584 transformed to unbound and bound B-chains:
Fig. 18-7: From
number 5´ in the ES-chain to B-chains in groups of 6:
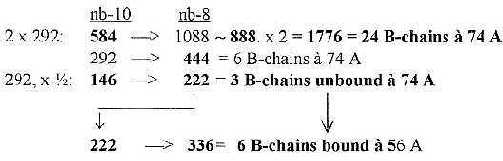
6.4 Total B-chains unbound times 2 from the 4 bases:
Fig. 18-8:
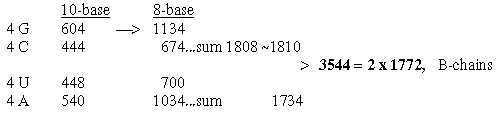
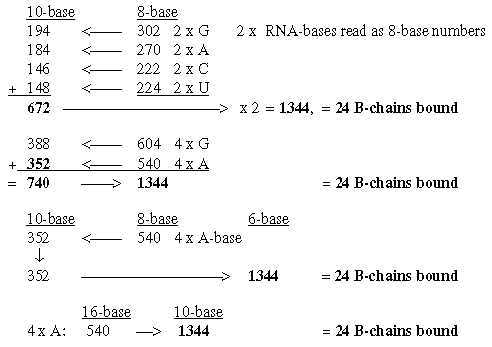
6.5 Total of bound B-chains = 1344 from the bases:
Fig. 18-9:
6.6 Hypoxanthine 136 in repeated steps gives B-chains bound or unbound:
Hypoxanthine 136 A (1/4 x 544) may give both B-chain
numbers 1344 and 1772 bound and unbound through 4 steps of transformations:
Fig. 18-10:

*Note that without rewritings 530 ~ 528 and 1020 ~ 1018 we get
1776 (24 x 74 A).
7. Displacements between 1st and 2nd base order: Numbers 220 - 26:
7.1 Relations between displacements 220 and 26:
Fig. 18-11:

Fig 18-12:
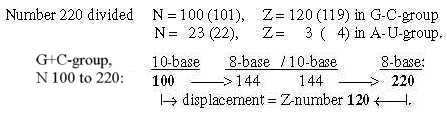
The relations between displacement 220 in the G+C-group and 26 in the U+A-group could be explained through only a minus 1 in N- and Z parts and the results in nb-8 through transformations.
Regard number 144 in figure 18-11 above divided in 64 and 80:
Fig. 18-13: How the displacement 220 and 26 could be explained through -1:
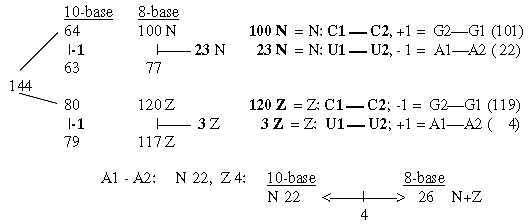
Fig. 18-14:
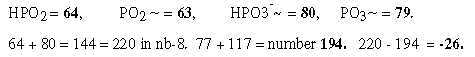
7.2 The number 220 in displacements in group G+C:
Fig. 18-15:

8. The 4 double-coded ams, sum = 246
The sum of R-chains of the 4 ams with two different codons are "also" 246,
i.e., the sum of displacements 220 and 26 above.
All 4 may become 37 in different nb-systems.
Fig. 18-16:
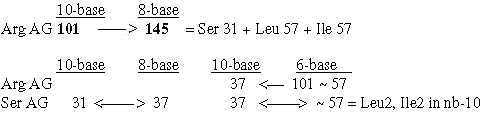
*
To P-groups