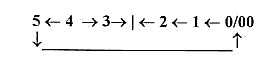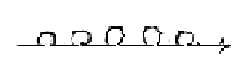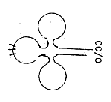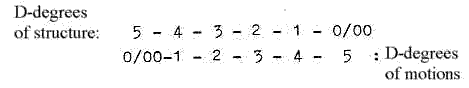From DNA to Amino Acids - the
Protein Synthesis: At
this very complicated level there
are some remarkable, simple features,
which seem to illustrate a dimension
chain of the kind suggested in the
pages about Physics: 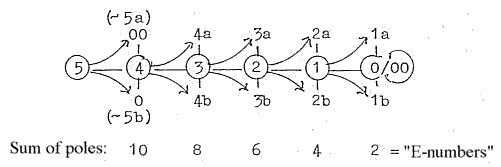
There are about 5 steps in the process (as in figure above of a
dimension chain on the model):
from
DNA →mRNA
→rRNA
→tRNA
→single
amino acid to proteins
(peptide
chains).
Forms in the process:
- mRNA as with a vector character,
4
- rRNA as "balls",
volumes, 3
- tRNA a surface structure: 2
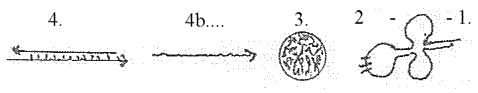
double
DNA-chains ----- mRNA ----- rRNA
--- tRNA ---- ams-----
|_______________________________________________________| 1.
We have the double DNA-spiral or
chain in the central nucleus of
the cell, a double-directed chain,
with bonds between complementary
code bases T--A, G--C as complementary
poles "0 - 00". (About
the complementary character see
here.)
2. This double chain gets polarized, the chains separate - and
one gets copied through contradictory direction from the manifold
of single code bases in the surrounding (as anticentre") to
mRNA, a single thread which moving outwards to the cytoplasm has
a vector character, (d-degree 4). It is directed to the ribosomes
located between the "centre and the anticentre" of the
cell, between the nucleus and cell wall.
3. rRNA,
the ribosomes, have the 3-dimensional
form of small spherical balls where
the mRNA gets attached.
Here
the meeting between tRNA and rRNA
occurs (at right angles, of natural
reasons, but compare the assumed
angle 90° in d-degree 3 in the
model). 4. tRNA, transferRNA,
are shorter chains of code bases
as nucleotides, chains separate
for separate amino acids, as each
lower dimension degree is a manifold
in relation to the higher d-degree.
tRNA has
the form of cloverleafs, partly
loops, partly linear, a 2-1-dimensional
figuration.
In the middle of
the central loop the codon for a
certain amino acid is located. 5.
In the opposite end of the tRNA,
at the linear end which is similar
in all tRNA molecules (bases A-C-C-),
the individual amino acid gets attached.
Then the
similar parts of the amino acids,
here called the "B-chains",
get bound through condensation to
protein chains, (in their turn then
forming 1-2-3-4-dimensional structures). The
5-dimensional chain can be understood
as double-directed from, 5→4→3
→
etc.
and "the other way
around" from 5 to 0/00 and
inwards.  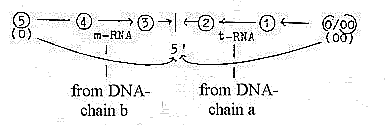
(1-dimensional
degrees branched off in steps outwards
transformed to motions in d-degree
0/00 and bound to 1-2-dimensional
forms meeting from anticentre (from
"outside", according to
description in Presentation of the
model.) Note: There are 5
histones, proteins of a certain,
fundamental kind, on which DNA is
rolled up: one of these 5 (called
H1 or H5) seems to have a function
related with this "all-embracing"
character of "the other way
around", linking DNA at both
ends of a spool of 8 histones, the
"nucleosome" (reference:
Wikipedia). (Each spool binds
146 base pairs of DNA: note the
number, 1/2 x 292: See page about
Numbers
or the pdf-file "Exponent
series".) Opposite
directions in the process and the
relation mRNA - tRNA:
The
difference between T- and U-base
represent the opposite directions:
- T inward direction towards
the DNA strand, when not active,
replaced by the
- U-base when
a DNA-strand is copied to mRNA
in the synthesis.
The only
difference is a CH3-group in the
T-base, obviously the chemical expression
for inward direction.
Many
bases in tRNA have got this group
added too, as if it were a fundamental
chemical sign for "inwards"
(+ 14 A). In the 4 examples
of tRNA given in the used reference
here,
there is also a T-base
in position 23 from the A-C-C-end,
a single one. Rather odd. And at
this position appears the same order
of the 5 bases:
T - U - C -
G - A.
The
ordinary U-bases of RNA is in tRNA
also turned (to "pseudouridine"),
as by an angle step, in their bond
to the ribose part of the nucleotides.
Compare the presumed angle steps
in the dimension chain. tRNA
with its 2-dimensional, partly wavelike
form (cf. suggested convex-concave
as geometrical poles of d-degree
2) and only 3 bases, the codon,
apparently essential for the protein
synthesis, has similarities with
the stage where mRNA on its way
to rRNA, undergoes change: irrelevant
parts of
its chain are cut
off. 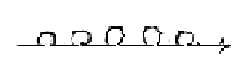 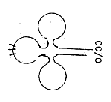
4
mRNA 3
2
tRNA 1
This
similarity could be read as a relation
between step 4 →
3 outwards and
2 1 inwards,
the dimension chain seen "orthogonal"
as double directed with a meeting
in step 3-2.
According
to the model we could have a 2-dimensional
motion resulting from step 4→3,
appearing inwards as a 2-dimensional
structure. 
(Chemists
have asked how it is possible for
the right ADP-transported amino
acid to find the right tRNA when
it is attached to the opposite end
of the tRNA in relation to the
place of its codon? If we presume
more of motions in d-degree 2 according
to the model, we could perhaps imagine
fast moments of neighbourhood between
these ends or "poles":
other stages of structure impossible
to detect in a laboratory ?) Why
this A-C-C-end of tRNA?
Similar
in all tRNA:
Triplets
of a dimension chain inwards and
mass numbers (A) of the bases: 012
111---
123....sum:
135 = A-base
111----
234....sum:
357 = A+C+C. Sum of the bases with
+1 for the bond to ribose.
↓
Intervals between the triplet numbers
= 111 = C-base, mass-number. 345,
the last triplet inwards, happens
to be the mass-number of the G-nucleotide
in chain bond, also = cGMP. GTP
(523 A) was, at least earlier, suspected
to take part in the peptide binding
of the amino acids.
rRNA
- the ribosomes: We
can note the locality of the small
spheres on the "endoplasmatic
reticulum", a structure of
membranes in cytoplasm - 3-dimensional
forms on 2-dimensional surfaces
and, as said above, in "the
middle" between the poles centre
- anticentre of the cell. It
is said (page 336 in the English
reference) that "groups of
5 or 6 individual ribosomes are
held together" by a molecule
of mRNA. Why just 5-6?
It's
said too that bacteria has 5 ribosomes!
We can also note that ribosomes
are made up of two parts with a
mass relation 2/1:
"50
S" and "30 S": 1,1
x 10-6 u and 0,55 x 10-6
u.
The parts unite through the
participation of Mg2+. Still
another thing about numbers:
Ribosomes consist of 40 - 60 % RNA,
60 - 40 % proteins, a 3-2- or 2-3
relation, as in the meeting in the
middle of the dimension chain between
the opposites:
code bases <-->
proteins.
Another
reference says the relation is 64
% RNA, 36 % protein:
that would
be a relation 8<2
- 6<2: compare
the E-numbers in d-degrees 3-2,
squared.
Number
of bindings in the synthesis:
- DNA - DNA, in the double chain:
a H-bond between atoms N ... H--O
- DNA - mRNA
- mRNA - rRNA
- tRNA - mRNA
- amino acid
- tRNA
- Amino acid - amino
acid: NH2---=O meeting,, condensation
(-H2O)
Number of
bases in the codons: 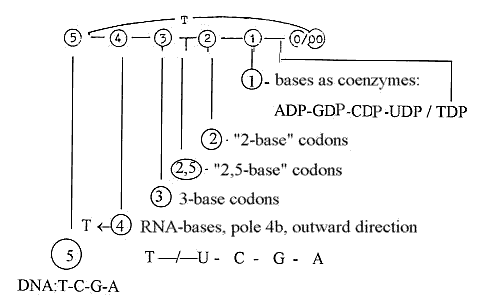
There
are 5 bases, T-U-C-G-A, getting
4 in DNA and mRNA, getting 3 in
the codons in the protein synthesis,
or 3 - 2,5 - 2:
the
3rd base indifferent in 1/3 if 24
codons and only separates A/G from
U/C-bases in 1/3 of the codons,
~"2,5" bases,
- getting
1 base in the role as coenzymes:
TTP-UTP-CTP-GTP-ATP
(or in
-DP-form as in the figure.)
Bases
- elementary kinds of substances: Carbohydrates
- Lipids - Proteins - Transportations
T
- U C
G
A T-
and U-bases in their role as coenzymes,
TTP and UTP, are used for bonds
in Carbohydrates (different kinds).
Note too
that all amino acids with U in first
or second position in their codons
are derived from the first stages
of the glycolysis where carbohydrates
are broken down. C as coenzyme
CTP is active in synthesis of Lipids. G
as coenzyme GTP takes part in the
Protein synthesis above. A
as coenzyme represent the central
energy store, as such corresponding
to a lot of transportation. A
dimension chain, according to the
model here, implies steps towards
growing amount of kinetic energy
(motions). Compare the similar
order of the bases in tRNA at position
23...: T-U-C-G-A!
(If the 4
examples should be typical.)
(Surely or presumably
the coenzymes of the bases take
part in many other special syntheses
and beak downs.) Circa
5 levels of storage of DNA:
Diameter (Ø) = 20 - 100 -
300 - 2000 - 6000 Å:
5 →
4: The double strand as helix, Ø
20 Å
4 →
3: This thread rolled up on balls
or cylinders of proteins
(histones?),
Ø 100 Å
3 →
2: These strand of balls drawn up
in a big spiral, Ø 300 Å
2 →
1: This in its turned drawn up in
a thicker spiral, Ø 2000
Å
1 →
0/00: In last step this thicker
spiral drawn up in a still
thicker
spiral, Ø 6000 Å,
making the arms
of the X-formed chromosomes,
and as a centre
of this the centromer, with a structure
of the type
0/00. Centre - anticentre,
~5' D-degrees should be viewed
as a suggestion. Alternatively we
could see the d-degree of motions,
from linear (1), 20 Å, to
rotation (2), on "balls",
100 Å, to spiral motions (3
dimensions), 300 Å, to spiralling
of higher orders in d-degrees 4
to 5, 2000 Å - 6000 Å.
Protein structures
1 - 2- -3 -4 -:
1 - Linear
chains,
2 - 2-dimensional surfaces
of laterally bound chains
3
- 3-dimensionally folded proteins,
globular proteins
4 - gathered
globular chains with centres of
coenzymes,
4
in haemoglobin for example. The
cilia or centromer structure perhaps
interpretable as a 5th level ?
DNA-bases - 5 bonds
between the two pairs: There
are 5 H-bonds (H for hydrogen) between
NH and O= , or NH and N divided
3-2:
3 between G- and C-base,
2 between A and U-base . Note
that G- and C-bases have both free
NH2- and =O groups,
while A-
and U-bases are "polarized",
A-base having only a free NH2-group,
U-base only =O-groups (keto-oxygen).
Some additional remarks:
Chromosomes
- "lumosomes":
- See
figure
about quotients between spectral
lines of the H-atom, centre in the
DNA double helix: Lyman-,
Balmer-, Paschen-series for spectral
lines of the H-atom: m = 2,
N = 5, 4, 3,→ &λ = 4341 - 4863 - 6565 Å: (1/4
- 1/25) / (1/4 - 1/16) = 112 x 10-2
112 = U-base l
(1/4 - 1/16)
/ (1/4 - 1/9 ) = 135 x 10-2 135
= A-base
(1/4 - 1/25) / (1/4
- 1/9 ) = 151,2 x10-2 151 = G-base And
what about C-base?
The lowest
spectral line of Oxygen 4368 Å.
Quotient to the the middle spectra
line here for H: 4863 Å =
111,3. x 10-2. (?)
[Quotients
between spectral lines could in
some way have relations to phase
waves.?] Could chromosomes
have some more fundamental property
common with light beams,
be
seen as "lumosomes" ?
Structured through the help of electromagnetic
waves?
What then should correspond
to the E and M factors in the EM-waves?
On the most fundamental
level perhaps the basic NHx-groups,
inwards, ~ M, versus the acidic
OHx- or =O-groups, outwards, ~E
?
On a secondary
level perhaps the chains of amino
acids as "radial" versus
codon bases
as "circular"
?
On a third
level the polarity between U-C-bases
and A-G-bases ? In any case,
the close relation between DNA/RNA
and proteins could get us presume
some kind of entity "DAP",
(DNA-Amino acids-Proteins), preceding
the development of the process,
according to views in this model.
An entity belonging totally to an
underground of mathematics?
To DNA-RNA-bases
- synthesis and complementarity
To Numbers
of Codon bases and some relations
to numbers of amino acids
*
HOME
|